The Bioinformatics/DNA search options allow you to search for amino acid strings and IUPAC codes in files containing DNA sequences. When one of these options is selected, the search string is treated as either a string of amino acid codes or IUPAC nucleotide codes.
Note that only exact matches are found. Imperfect matches (as in BLAST searches) are not supported.
By default, these new options are hidden from the user. To enable the new functionality, you must enable the BioInformatics/DNA search option in the Search section of Preferences.
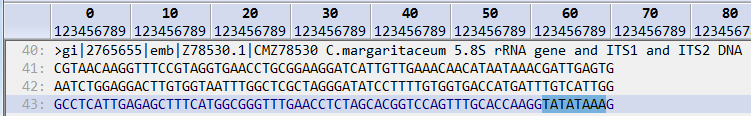
There are 20 valid codes which represent the 20 standard amino acids (ACDEFGHIKLMNPQRSTVWY). An X is used to indicate any amino acid and a * is used to indicate a stop codon.
V will search the file (containing DNA sequences) and will match any sequence that corresponds to the entered amino acid string.
The screenshot below shows the result of searching for an amino acid string of GRGDS
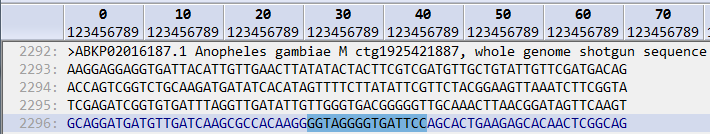
There are 15 valid codes which correspond to the valid characters in the DNA/RNA alphabet (ACGTURYMKSYNDHV), and N is used to indicate any nucleotide.
The screenshot below shows the result of searching for an IUPAC nucletode string of TATAWAAM